Scientists empower the computational design of biotherapeutics
Anton Bushuiev, Roman Bushuiev as well as several other researchers from the team of Josef Šivic at CIIRC CTU, in collaboration with the team of Stanislav Mazurenko at Loschmidt Laboratories at Masaryk University and International Clinical Research Centre (ICRC Brno), and the team of Tomáš Pluskal at IOCB Prague, have developed a new method to improve the computational design of biotherapeutics. Their new machine-learning method enables more efficient design of proteins with better interaction properties. The results will be presented at one of the world’s top machine learning conferences – ICLR 2024 in Vienna.
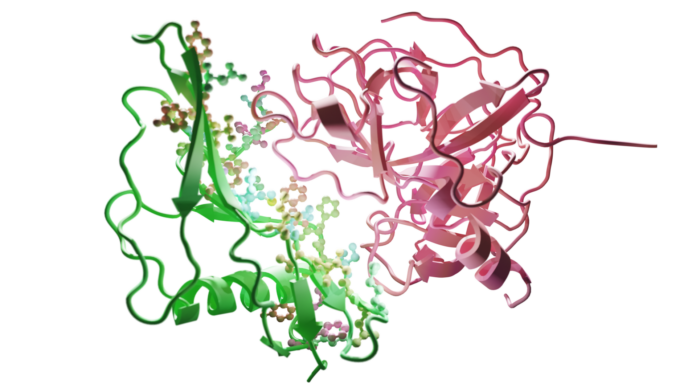
Interaction of the staphylokinase-based drug candidate (left/green) with the plasmin protein (right/red) present in human blood. The amino acids of staphylokinase responsible for the interaction are shown by small spheres in different colors.
he motivation is the design of an improved version of the staphylokinase protein, an attractive thrombolytic drug candidate for breaking down blood clots during stroke. Strokes can have serious and long-lasting effects, including paralysis, speech and language problems, memory loss, and emotional difficulties. The impact can vary widely depending on the severity of the stroke and how quickly treatment is received. The widespread clinical application of staphylokinase is currently limited due to its weak interaction with the plasmin protein present in human blood.
„We aim to improve staphylokinase by replacing some of its building blocks – the amino acids – responsible for the interaction. This requires discovering amino-acid substitutions that enhance the desired interaction properties out of millions of possible candidates“, explains principal investigator Josef Sivic from CIIRC CTU. Identification of such amino acid substitutions is of high practical interest also for other tasks involving the interaction of proteins such as the design of vaccines and biosensors.
The proposed method called PPIformer, described in the paper „Learning to design protein-protein interactions with enhanced generalization“, enables predicting the effects of amino acid substitutions on protein-protein interactions (PPIs) in a fraction of a second. At the core of PPIformer is a machine learning approach that implements so-called „self-supervised“ learning that does not require costly and slow laboratory experiments. Instead, it relies on the newly collected and currently the largest dataset of unannotated protein-protein interactions, mined from all publicly known protein structures.
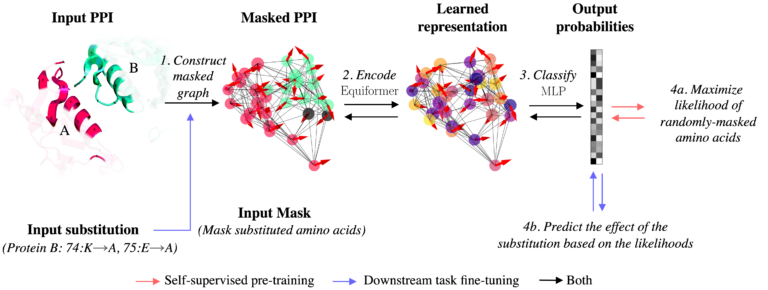
The illustration of the proposed method that takes the protein interaction together with a target substitution as input (left) and predicts the effect of the substitution based on the predicted probabilities of possible amino acids (right).
PPIformer was first trained to accurately identify amino acids masked in the protein-protein interaction structures, like how the current large-scale language models, such as ChatGPT, are trained by predicting masked words in natural language sentences.
„After learning from millions of such self-supervised masked amino acid training examples, the trained neural network model was adapted to predict the effects of amino-acid substitutions via a short fine-tuning on scarce in-laboratory measured data,“ says doctoral student and main author of the method Anthon Bushuiev. The developed method has demonstrated high potential in identifying favourable mutations in staphylokinase, as well as in a human antibody against coronavirus.
The designs of improved staphylokinase found by the proposed approach are currently being experimentally validated at Loschmidt Laboratories at Masaryk University and the International Clinical Research Center in Brno.
Next, the team is looking into extending this approach to biomolecules involved in neurodegeneration in collaboration with the International Neurodegenerative Disorders Research Center (INDRC).
The developed machine learning method was accepted to ICLR 2024 – The Twelfth International Conference on Learning Representations. ICLR is one of the most influential conferences in machine learning (along with NeurIPS and ICML) and is ranked among the top ten most influential journals and conferences in all areas of science by Google Scholar.
Original article: A. Bushuiev, R. Bushuiev, P. Kouba, A. Filkin, M. Gabrielova, M. Gabriel, J. Sedlar, T. Pluskal, J. Damborsky, S. Mazurenko, J. Sivic, Learning to design protein-protein interactions with enhanced generalization, International Conference on Learning Representations (ICLR), 2024. https://doi.org/10.48550/arXiv.2310.18515

